CoV-ncRNASig
![]()
Motivation. Due to viral infection, the human immune system activates a complex response whose magnitude also depends on the interplay between the virus and the regulation of the host’s immune response. The most dangerous effects induced by the SARS-CoV2 infection are an exacerbated inflammatory response and an extensive lung pathology. Related to the damages caused by the inflammatory response, an important aspect that deserves to be investigated is the host cell response at a very early stage of the virus infection. An extensive analysis of transcriptome profiles of infected cells is the most effective analysis approach to investigate at what extent and which gene signalling pathways are directly involved at this stage. Elucidating these aspects can lead to the identification of biomarkers of infection and targets for new and more effective therapeutic approaches.
It is noteworthy that non-coding RNAs (ncRNAs) are essential regulators of human gene expression. Recent studies have demonstrated that viruses belonging to the family of SARS-COV-2 can regulate the expression of small (sRNA) and long non-coding RNAs (lncRNA) [1,2]. We have investigated the potential of the SARS-COV-2 genome transcription to produce fragments of RNAs that can interfere with the host regulatory non-coding RNAs (small non-coding RNAs) by using a large scale bioinformatics analysis on data available in public repositories.
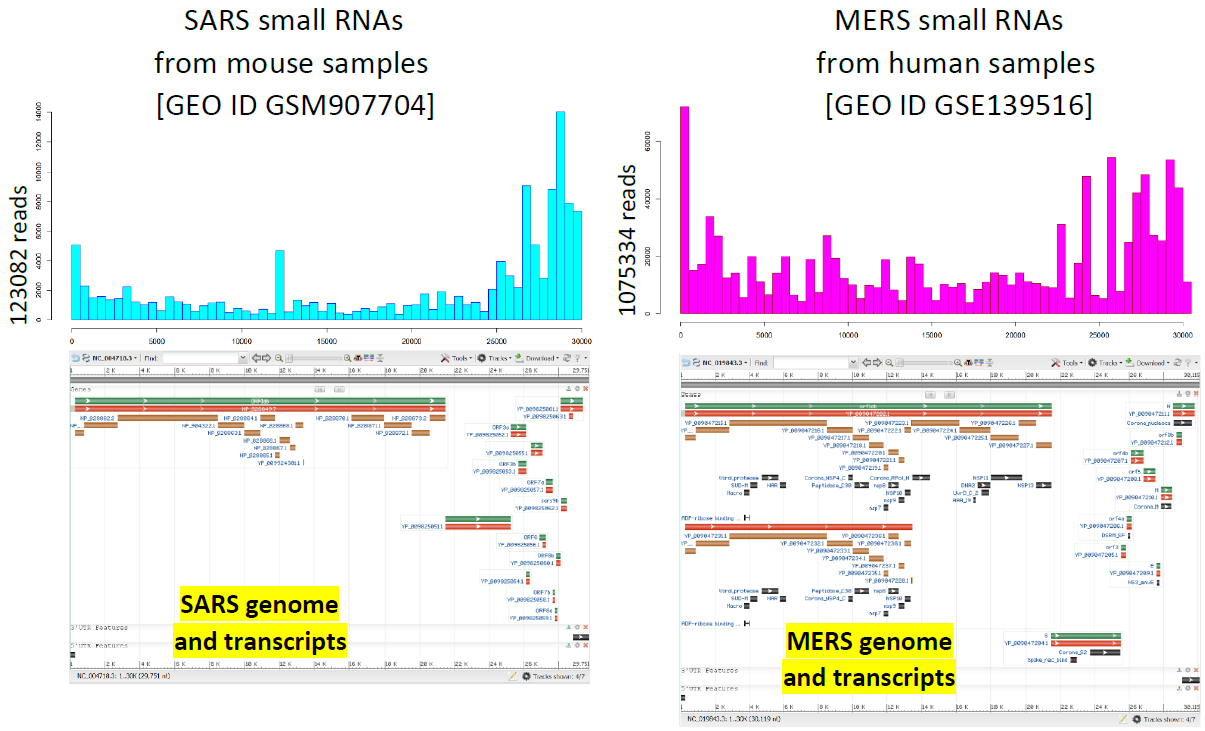
Results. Comparing the SARS-CoV2 genome sequence (NC_045512.2) with RNA-seq data of human lung cancer cells infected with MERS-CoV [GEO ID GSE139516], mouse lung cells affected by SARS-CoV [GEO ID GSM907704] and bronchial lavages and Peripheral Blood Mononuclear Cells (PBMC) from COVID-19 patients [3] we discovered that small fragments of ncRNAs from SARS-CoV2 might interfere with the activity of endogenous miRNAs that target genes involved in the inflammatory response and in particular with the allergic asthmatic reaction (IL4-IL13 signalling pathway).
These preliminary results open the way for more effective treatment of COVID-19 patients and defence from future coronavirus pandemics.
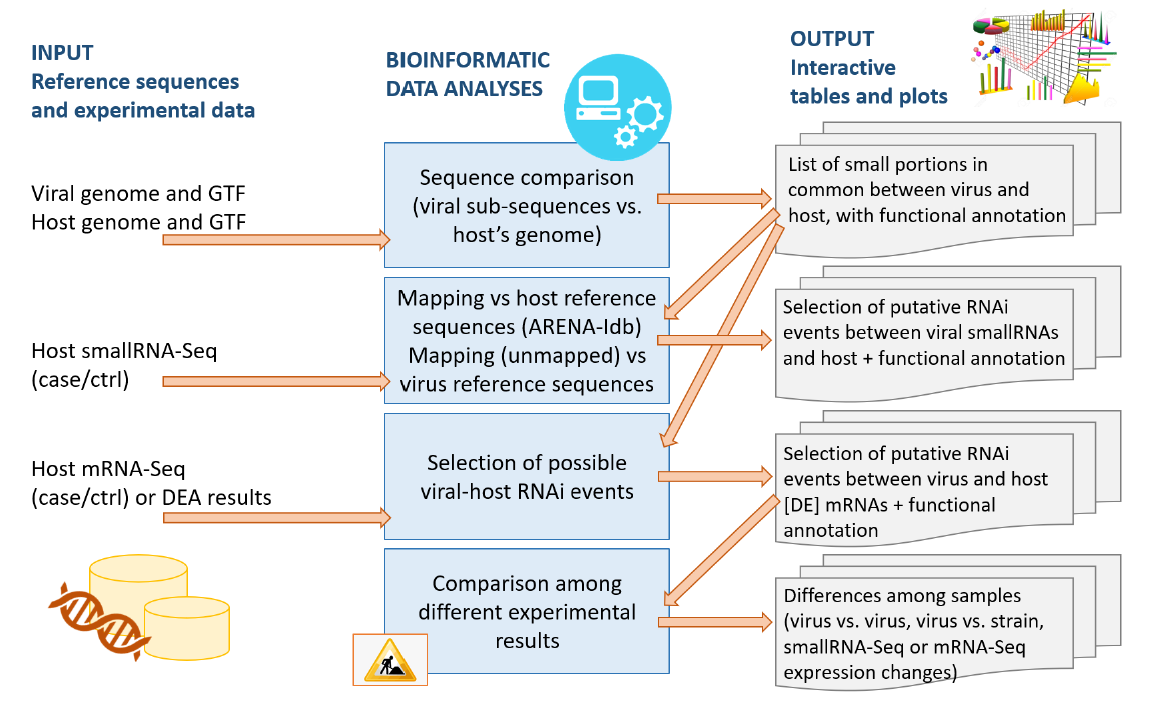
Methods. The analysis was carried out with a bioinformatic pipeline developed using Python, BASH and R, the software BLAST, STAR and mirdeep2 for the comparative analyses and a database of small non-coding RNAs (developed by our Bioinformatics group), named “Arena-Idb” [4], as reference database. The pipeline that we call CoV-ncRNASig, is composed by three analysis modules.
The first module takes as input the viral genome and the genome of the host, and compares them in search of common sequences that are at least 18 nucleotides long. This comparison is performed with a pipeline developed ad-hoc that includes multiple strategies, starting from a simple sequence comparison (with a maximum of 2 mismatches allowed), and continuing with functional binding predictions such as micro RNA seed-oriented pairing. The result is a list of common sub-sequences, containing both the viral and the host annotations.
The second module searches for small viral RNAs in sequencing data of the host. We compare to the viral genome the reads that do not match with human genome and transcriptome. Then we cross-correlate such results with the genome analysis results by searching for those viral RNAs that partially match the host sub-sequences. The output is the list of annotated sequences and molecular pathways of the host that should suffer from the viral small RNA interference. If for the same host, also NGS transcriptome data are present, we also search for differentially expressed transcripts whose expression could be influenced by the viral RNA interference action.
The last module allows the user to integrates the results of different executions of the first two modules. This module highlights similarities and differences between the analyses performed on different samples to compare (for example) the effect of different viruses or of different strains of the same virus on the same host, or to study the potential impact of mutations in the infection process, in both, the viral and the host’s genome.
Further information and updates will be published at http://bioinformatics.ba.itb.cnr.it/CoV-ncRNASig
References:
[1] Morales et al., (2017), SARS-CoV-Encoded Small RNAs Contribute to Infection-Associated Lung Pathology, Cell Host & Microbe 21, 344–55. doi: 10.1016/j.chom.2017.01.015
[2] Weiwei Liu and Chan Ding, (2017), Roles of LncRNAs in Viral Infections. Front Cell Infect Microbiol. 2017; 7: 205. doi: 10.3389/fcimb.2017.00205
[3] Yong Xiong et al. (2020), Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerging Microbes & Infections, 9(1):761-770. doi: 10.1080/22221751.2020.1747363
[4] Bonnici et al. (2018), Arena-Idb: a platform to build human non-coding RNA interaction networks. BMC Bioinformatics 2018 19(Suppl 10):2298. doi: 10.1186/s12859-018-2298-8
Keywords: non-codingRNA, RNA-interference, host-virus-interaction
Contacts: arianna.consiglio | domenico.catalano | giorgio.grillo | flavio.licciulli | domenica.delia + @ba.itb.cnr.it
Authors: Arianna Consiglio, Domenico Catalano, Giorgio Grillo, Flavio Licciulli, Domenica D’Elia
Bioinformatics Facilities @ ITB Bari | Powered by CMSimple| Template: ge-webdesign.de| Login